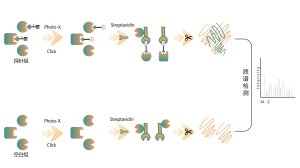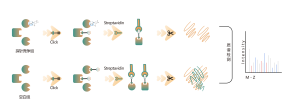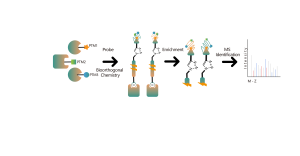
Products
Competitive Chemoproteomic Profiling of Protein Targets for Covalent Small Molecule Drugs
Platform Technical Features
Similar to non-covalent drugs, the direct pulldown strategy by using chemical probes functionalized with reporter groups have also been demonstrated successfully for covalent small molecule drugs. However, it is worth noting that some covalent drugs are intolerant of chemical modifications due to loss of bioactivity or synthetic inaccessibility. In addition, the formed covalent bond is usually unstable during MS detection.
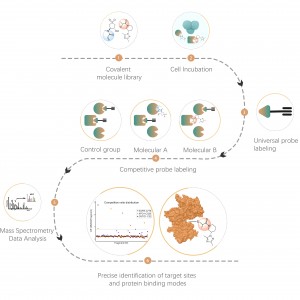
Competitive chemoproteomic strategy is an ideal alternative, which uses a universal activity-based probe for protein labeling. Specific chemical probes have developed to react with cysteine, lysine, serine, and histidine residues. In principle, once occupied by a covalent small molecule, the amino acid residue cannot be labelled by the probe. As a result, the ON and OFF-targets could be identified comprehensively by this competitive strategy with the resolution of amino acid.

Workflow

The platform follows a structured workflow, starting with living cell labeling with covalent molecules, followed by steps include labeled proteome extraction, chemical probe labeling, bioorthogonal ligation, streptavidin-based enrichment, protease digestion, isotopic labeling, and final mass spectrometry detection.
Case Study
Project Aim
AMG510, developed by Amgen, is the first approved drug targeting KRAS-G12C mutation in tumors. The aim of the project was to profile the on and off targets of AMG510 in NCI-H358 cells with the KRAS-G12C mutation.

Experimental Method
DIA-ABPP, one of the core technology platforms in ChomiX, was used to analyze the target occupancy in the proteome level.
Data Visualization



A total of 16992 Cys sites from 5768 proteins were quantified in NCI-H358 cells. The target occupancy of KRAS_C12 site was close to 100% under the treatment of 1 μM AMG510 in situ, while the other site, KRAS_C80, was unaffected. This data demonstrated the high target specificity of AMG510. (The labeled Cys residues are indicated with *)