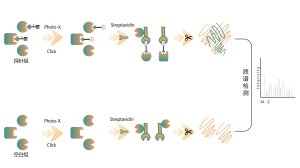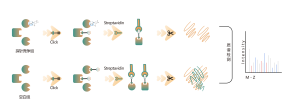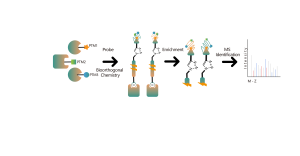
Products
Chemoproteomic Profiling of Protein Targets for Noncovalent Small Molecule Drugs
Platform Technical Features
Small molecule drugs play an important role in the field of drug R&D. The current FDA approved drugs target a total of 812 distinct human proteins. Among the drugs directed against the above-mentioned targets, 84% are small molecule drugs. Moreover, only 639 of these proteins have been targeted with small molecule drugs. The interaction between small molecule drug and protein target includes non-covalent and covalent modes, and the former is currently dominant.
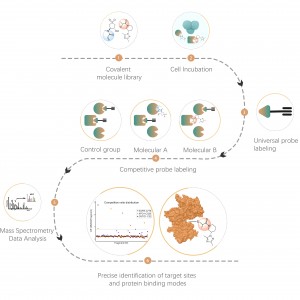
Non-covalent interactions, such as hydrogen bonds and π-π stacking, can be disrupted due to protein denaturation. To address this challenge, our platform employs photoaffinity labeling, a well-established technique for precisely attaching "chemical labels" to a protein's active site. Furthermore, our innovative in situ chemical crosslinking strategy transforms transient non-covalent protein interactions into covalent and permanent chemical bonds. By utilizing a chemical probe that is functionalized with both photoaffinity and bioorthogonal moieties, ChomiX's chemoproteomics platform has demonstrated its effectiveness in successfully fishing out protein targets within cell lysates, tissues, and living cells. The spectrum of bioactive small molecule drugs applied in the platform encompasses a variety of compounds, including endogenous metabolites, natural products, and non-covalent synthetic molecules.

Workflow

The platform follows a structured workflow, starting with the labeling of living cells using a photoaffinity probe derived from non-covalent molecules. Subsequent steps include the extraction of labeled proteomes, bioorthogonal ligation, streptavidin-based enrichment, protease digestion, isotopic labeling, and finally, mass spectrometry detection.
Case Study
Project Aim
Compound A showed good anti-proliferation activity in the cell viability assay. Chemoproteomics platform was used to characterize the protein targets.
Experimental Method
The photoaffinity chemical probe, Probe A, was designed and synthesized based on the SAR data of compound A. Probe A also showed similar anti-proliferation activity in the tumor cell line. Gel and MS-based experiments were performed. The MS data were analyzed for the elucidation of MOA.
Data Visualization

The gel-based fluorescence results showed that Probe A could effectively label proteins, and the labeling signal could be significantly competed by compound A. Collectively, these data indicated that Probe A was able to be used as the chemical probe tool for subsequent target discovery since it could bind the same proteins as compound A.



Volcano plot showed that 114 proteins (red highlight) were enriched by Probe A significantly in the Probe A vs DMSO (direct) group, and 38 proteins (red highlight) were competed by compound A significantly in the Probe A vs (A+Probe A) (Competition) group. Venn diagrams showed that 32 overlapped proteins could be potential compound A-binding targets with high confidence. (n = 3, ratio ≥2,p-value ≤ 0.05)