Technischer Hintergrund
Die chemische Proteomiktechnologie erweist sich als ein Leuchtturm des Wandels, der die Arzneimittelforschung weg von gereinigten Proteinen und hin zum Bereich lebender Zellen lenkt.
Die chemische Proteomiktechnologie dient als robuste Plattform zur Arzneimittelentdeckung und ist durch die Untersuchung kleiner Molekül-Protein-Wechselwirkungen in lebenden Zellen gekennzeichnet. Eine unserer kovalenten Arzneimittelforschungsplattformen ist darauf ausgelegt, kovalente Bindemittel zu entdecken, die Proteinziele durch kovalente Reaktionen mit der Thiolgruppe von Cysteinresten angreifen. Im Gegensatz zu herkömmlichen Single-Target-Screening-Strategien ermöglicht die chemische Proteomik-Plattform eine quantitative Analyse der Wechselwirkungen zwischen elektrophilen Fragmenten und nahezu allen Proteinen in lebenden Zellen mit der Auflösung von Aminosäureresten. Es bietet eine Abdeckung von mehr als 10.000 Proteinen und~40.000 Cysteinstellen (potenzielle Arzneimittelbindungsstellen) aus mehreren Säugetierzelllinien.

Arbeitsablauf
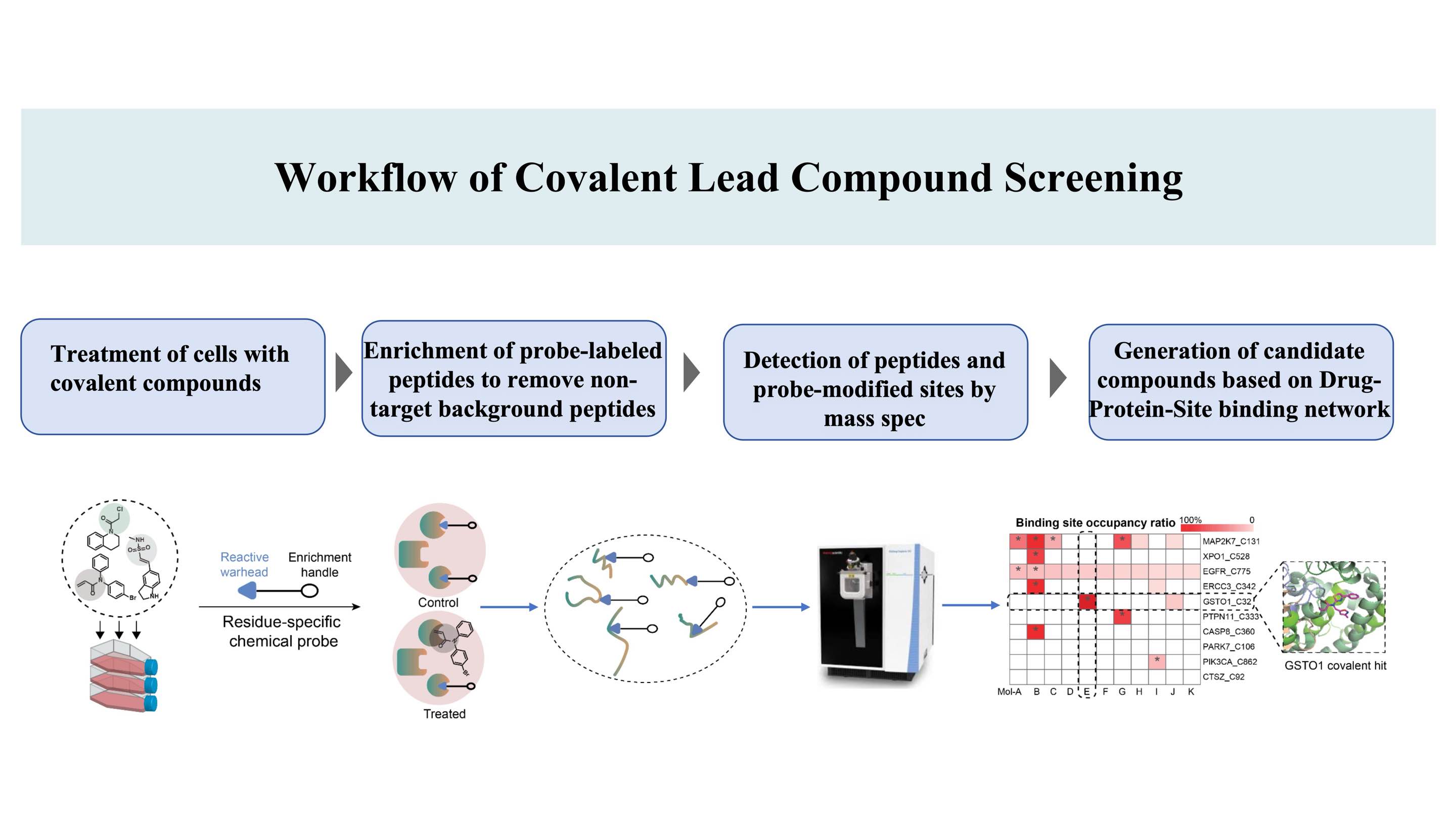
Der Arbeitsablauf zur Entdeckung kovalenter Verbindungen, die auf Bindungsstellen* auf Proteinen abzielen, basiert auf dem DIA-ABPP-Patent (Data-Independent Acquisition-Activity-Based Protein Profiling) (Eine kovalente Bindungsstelle ist eine Aminosäure, die durch chemische Sonden markiert werden kann, wodurch die Ligandierbarkeit ermöglicht wird)
Technische Vorteile
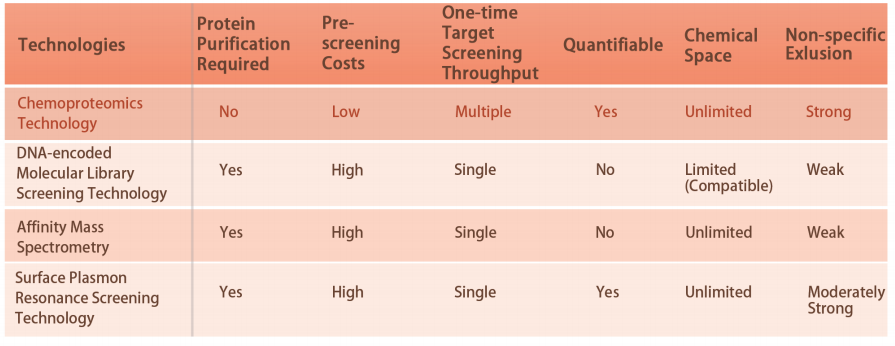
Merkmal 1
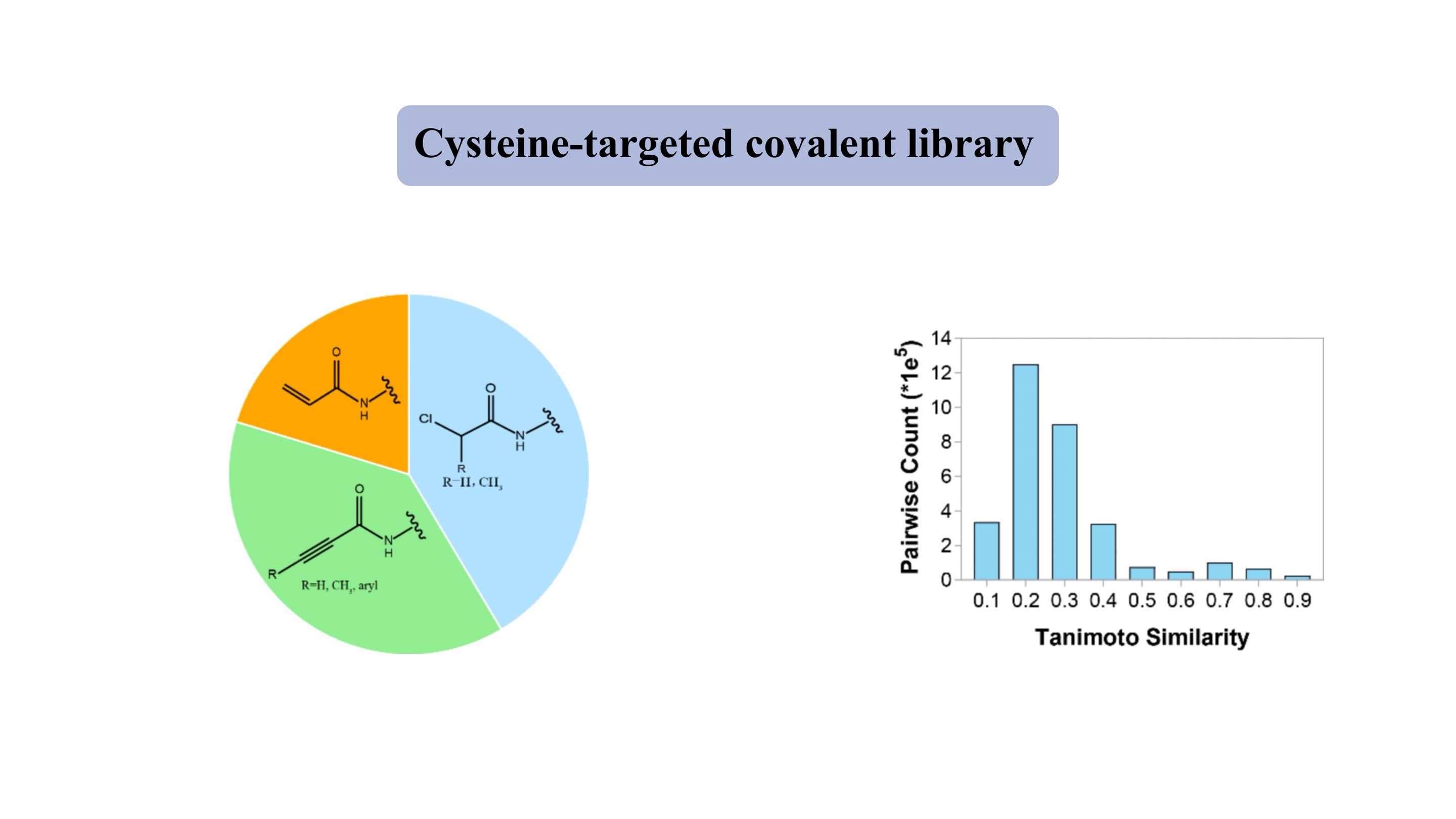
Auf Cystein ausgerichtete kovalente Bibliothek
Die auf Cystein ausgerichtete kovalente Bibliothek enthält repräsentative milde elektrophile „Gefechtsköpfe“ wie Acrylamide und Chloracetamide. Die „wirkstoffähnliche“ Bibliothek enthält etwa 3000 Verbindungen, von denen mehr als 80 % ein Molekulargewicht von 300–500 Da aufweisen. Bei den meisten Verbindungen liegt der Tanimoto-Ähnlichkeitsindex bei etwa 0,3 für jeweils zwei Mitglieder, was auf einen hohen Grad an Diversität hinweist.
Merkmal 2
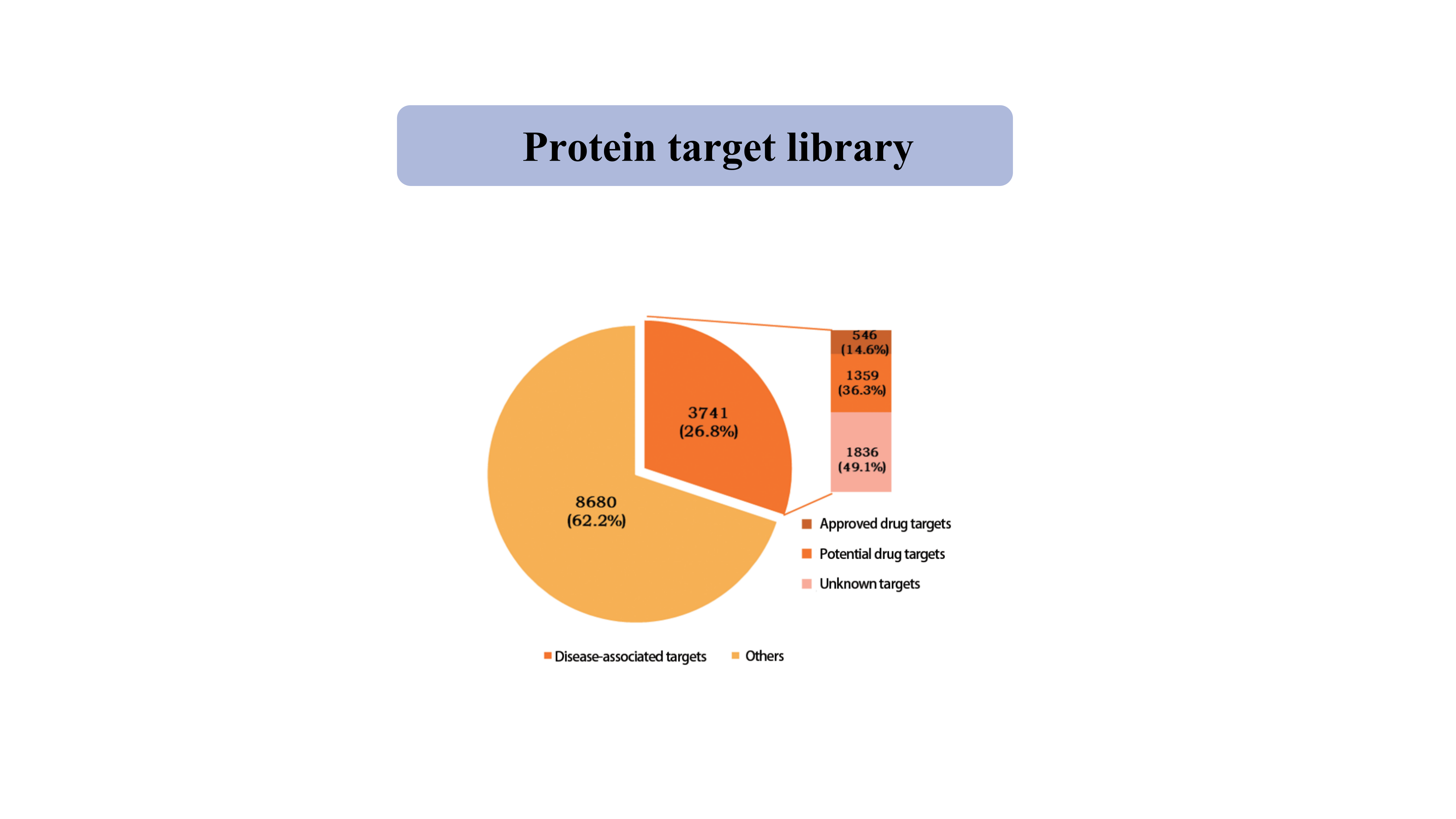
ProteinzielBibliothek
Derzeit umfasst die Bibliothek der von der Thiol-spezifischen chemischen Sonde erfassten Proteinziele 39.962 Cysteinstellen von 12.421 Proteinen, einschließlich Kinase, Phosphatase, Ligasen und Transkriptionsfaktoren.
Fallstudie
DCAF1 dient als Substratrezeptor für zwei unterschiedliche E3-Ligasen (CRL4DCAF1 und EDVP) und spielt eine entscheidende physiologische Rolle beim Proteinabbau. Mehrere kovalente und nichtkovalente Bindemittel, die auf die WDR-Domäne von DCAF1 abzielen, wurden entwickelt, um gezielte Abbauanwendungen zu unterstützen (Targeted Protein Degradation by Electrophilic PROTACs that Stereoselectively and Site-Specifically Engage DCAF1. J. Am. Chem. Soc. 2022, 144, 40, 18688). –18699. DCAF1-basierte PROTACs mit Aktivität gegen klinisch validierte Ziele zur Überwindung von intrinsischem und erworbenem Abbauwiderstand (Nat. 2024, 15, 275).
Wir haben zum ersten Mal entdeckt, dass Mol-380 kovalent mit DCAF1_C69 interagiert, was es als potenzielle medikamentöse Stelle für TPD-Anwendungen unabhängig von der WDR-Domäne hervorhebt. Unsere Ergebnisse unterstreichen den erheblichen Wert der automatisierten chemischen Proteomikplattform ChomiX bei der Entdeckung neuer Liganden für nicht behandelbare Ziele in lebenden Zellen, einschließlich Transkriptionsfaktoren und Membranproteinen, und unterstreichen ihren potenziellen Einfluss auf die Arzneimittelentwicklung und funktionelle Erforschung.
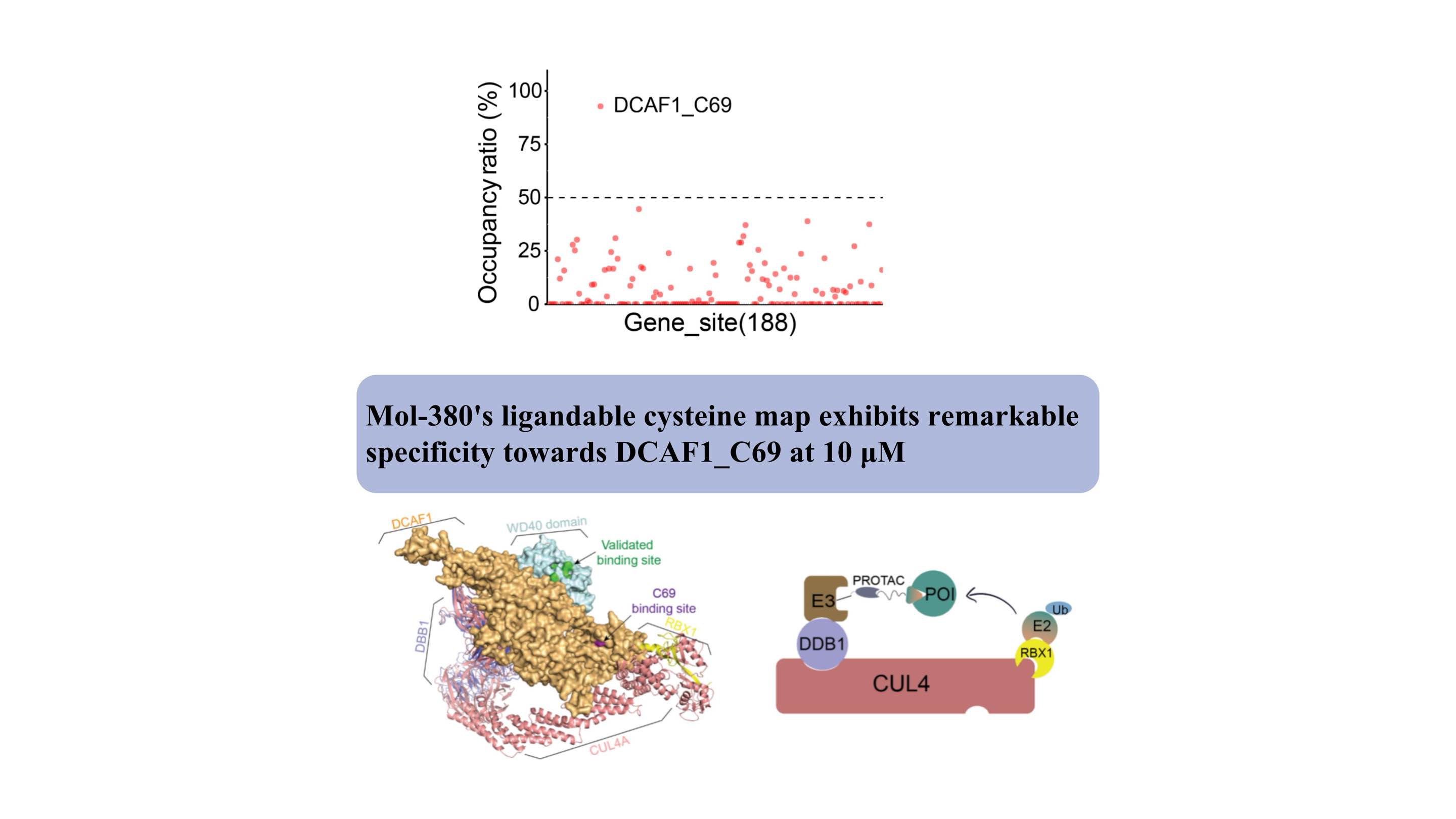
Strukturell liegt die Bindungsstelle von C69 neben der validierten Tasche der WD40-Domäne, wie das komplexe Modell zeigt, und bietet eine neue Stelle für die Entwicklung von PROTACs.