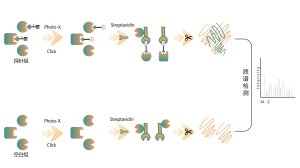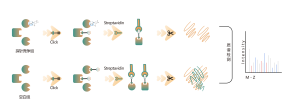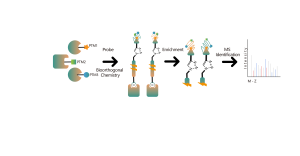
Products
Target Identification and Selectivity Analysis of Targeted Protein Degraders
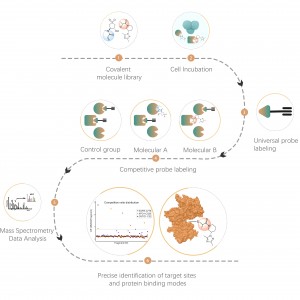
The Proteolysis Targeting Chimeras (PROTAC) drug is a bifunctional molecule that can bind to the targeted protein and recruit the ubiquitin E3 ligase for degradation. Therefore, novel modalities such as PROTAC and molecular glue have the ability to inadvertently alter the abundance of endogenous proteins, which provides a new therapeutic method for protein targets, especially the undruggable proteins. Comprehensively quantifying the abundance of on-target and off-target proteins has been one of the standard experiments for TPD drugs R&D.

Technical Platform
ChomiX has developed innovative quantitative chemical proteomics platforms, allowing precise analysis of over 5,000 proteins in a single cell line, supporting comprehensive target selectivity studies. For example, the Tandem Mass Tag (TMT) platform extracts and processes proteomes from cells treated with protein degraders, using enzymatic digestion, labeling, and liquid chromatography for deep proteome coverage. Mass spectrometry then analyzes protein abundance differences, providing critical target and selectivity data for protein degrader development. Additionally, ChomiX’s Data Independent Acquisition (DIA) platform efficiently quantifies low-abundance proteins in complex samples, identifying subtle differences across the proteome.

Our Advantages
1. Technical Excellence: Experienced team, top-tier journal publications, and authoritative industry services.
2. Core Patent Technology: Exclusive patents and advanced hardware for early drug development support.
3. One-stop Service: Covering probe design, synthesis, target discovery, bioinformatics analysis, and timely progress feedback for customer satisfaction.
4. Rigorous Quality Management: ISO9001 certification ensures trustworthy and authentic reports.
Our Service
| Project | Identification and Selectivity Analysis of Protein Degrader Targets |
| Sample | Cell lysate, live cells |
| Hardware Platform | Non-contact ultrasonic cell pulverizer,ChemiDoc MP Imaging System,Orbitrap Fusion Lumos Tribrid/Orbitrap Exploris 480/Q Exactive HF-X/timsTOF Pro 2 mass spectrometer |
| Project Duration | 3-4 weeks |
| Deliverables | Project Report (including experimental procedures, data analysis charts, bioinformatics analysis results) |
| Price | Click to consult |
Case Study
Project aim: Two PROTAC molecules were designed and synthesized. The on and off-target proteins need to be analyzed for comparison of selectivity.
Experimental method: TMT-based quantitative MS method was used for proteomic analysis.
Project results:

A total of 5900 proteins were quantified in tumor cell lines and ON & OFF targets were identified for PROTAC1 and 2 molecules (Orbitrap Exploris 480 mass spectrometer, TMT10plex and data analysis by PD 2.5)