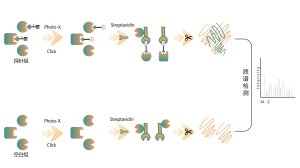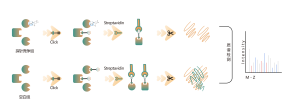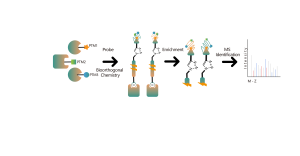
Products
Identification of non-covalent small molecule drug targets based on photoaffinity probes
Technical Platform
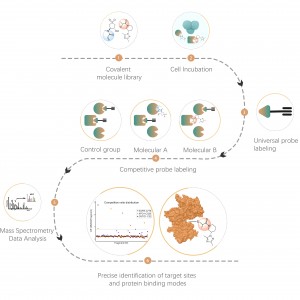
The chemical proteomics target identification platform based on photoaffinity probes involves several key steps, including probe design, synthesis, activity assessment, labeling, protein enrichment, and data analysis. Non-covalent small molecule drugs, such as synthetic compounds, herbal extracts, natural products, and metabolites, can be modified into photoaffinity probes. Once these probes bind to their targets within cells, they form stable covalent interactions, allowing for the selective enrichment and identification of low-abundance target proteins.

Our Advantages
1. Technical Excellence: Experienced team, top-tier journal publications, and authoritative industry services.
2. Core Patent Technology: Exclusive patents and advanced hardware for early drug development support.
3. One-stop Service: Covering probe design, synthesis, target discovery, bioinformatics analysis, and timely progress feedback for customer satisfaction.
4. Rigorous Quality Management: ISO9001 certification ensures trustworthy and authentic reports.
Our Service
| Project | Identification of Direct Targets for Non-covalent Small Molecule Drugs |
| Sample | Recombinant protein, cell lysate, live cells, diseased tissue, blood, bacteria, plant tissue |
| Hardware Platform | Non-contact ultrasonic cell pulverizer,ChemiDoc MP Imaging System,Orbitrap Fusion Lumos Tribrid/Orbitrap Exploris 480/Q Exactive HF-X/timsTOF Pro 2 mass spectrometer |
| Project Duration | 4-8 weeks |
| Deliverables | Project Report (including experimental procedures, data analysis charts, bioinformatics analysis results) |
Case Study
During the drug screening process, utilizing cell viability screening technology, compound A was found to exhibit significant inhibitory effects on target cells. To further identify its target proteins at the molecular level, decipher its mechanism of action, and explore potential new targets, our company designed and synthesized photoreactive probe Probe A (incorporating photoreactive and bioorthogonal groups) based on the structure and activity characteristics of compound A. Leveraging the chemical proteomics technology platform, we employed fluorescence labeling and mass spectrometry techniques for target protein identification in cell lines relevant to the activity. Combined with bioinformatics analysis methods, we delved deeper into the mechanism of action of compound A and its associated novel target proteins.

Based on fluorescence gel analysis of the labeling experiment, Probe A effectively labels proteins, and the labeling signal can be significantly competed by compound A. This indicates that Probe A has a similar target coverage to compound A, making it a suitable chemical probe tool for subsequent target discovery.




The Volcano plot illustrates the results of the Probe A vs DMSO (Direct) experiment, where 114 proteins (highlighted in red in the upper plot) were significantly enriched by Probe A. In the Probe A vs (A+Probe A) (Competition) experiment, 38 proteins (highlighted in red in the lower plot) were labeled by Probe A and significantly competed by the original compound A. These two experiments generated 32 proteins with high confidence binding to compound A (n = 3, ratio ≥ 2, p-value ≤ 0.05). GO Biological Pathway analysis of the 32 proteins with high confidence binding to compound A revealed significant enrichment in signaling pathways such as phospholipid efflux, negative regulation of lipase activity, and regulation of sterol transport, aligning with the phenotype.