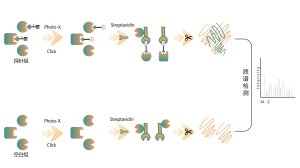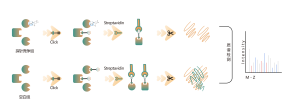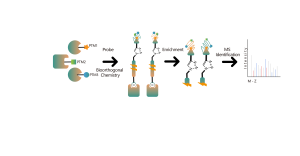
Products
Identification of Protein-Protein, Antibody-Antigen Interaction Regions: Chemical Cross-Linking
Understanding protein structure and interactions is essential for uncovering their biological functions. Due to the proteome’s complexity, no single technique can fully reveal these aspects. Biologists often use a combination of methods, with cross-linking mass spectrometry (XL-MS) standing out for its unique advantages.
XL-MS provides precise spatial distance information between amino acid residues and offers numerous benefits: minimal sample requirement, low environmental demands, no molecular weight limitations, in situ cross-linking, ease of use, and extensive data output.
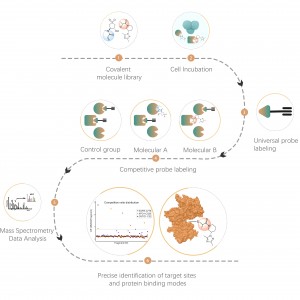
ChomiX's XL-MS technology uses various chemical cross-linkers to target specific amino acid residues, precisely cross-linking and analyzing protein-protein and antibody-antigen interactions.
Technical Platform
Common chemical cross-linkers (as shown in the figure below) can covalently cross-link specific amino acid residues when added to protein systems. After enzymatic digestion, cross-linked peptides are generated, representing the interaction regions between proteins. Through mass spectrometry identification and precise analysis of the peptide sequences, information about the interaction regions can be obtained.

Amino-to-thiol cross-linker BS3

Amino-sulfhydryl crosslinker EMCS

Our Advantages
1. Professional Excellence: Our team boasts extensive experience and publications in top journals, offering industry-leading technical services.
2. Efficient Solutions: We employ reliable methods to drive projects forward swiftly, providing worry-free solutions.
3. Rigorous Quality Management: Adhering to ISO 9001 standards, our mature quality management system ensures the authenticity and reliability of our reports.
4. Systematic Project Management: From consultation to report delivery, we provide timely progress updates, ensuring customer satisfaction and efficient project execution.
5. Cutting-Edge Equipment: Equipped with advanced mass spectrometers like the Thermo Fisher Orbitrap Exploris 480 and Bruker timsTOF, we facilitate groundbreaking research.
Our Service
| Project | Identification of interaction regions for protein-protein and antibody-antigen interactions |
| Sample | Pure protein, antibody-antigen complex |
| Hardware Platform | Non-contact ultrasonic cell pulverizer,ChemiDoc MP Imaging System,Orbitrap Fusion Lumos Tribrid/Orbitrap Exploris 480/Q Exactive HF-X/timsTOF Pro 2 mass spectrometer |
| Project Duration | 2-4 weeks |
| Deliverables | Project Report (including experimental procedures, data analysis charts, bioinformatics analysis results) |
| Price | Click to consult |
Case Study

In this case, BSA serum protein was cross-linked using the amine-reactive cross-linker BS3 to construct a dimeric structure of BSA serum protein. Mass spectrometry analysis of the dimeric structure identified cross-linked peptide segments that matched existing literature data, thereby validating the accuracy and reliability of the experiment.

The cross-linked peptides actually detected are consistent with the cross-linked peptides presented in the following literature- Characterization of protein unfolding by fast cross-linking mass spectrometry using di-ortho-phthalaldehyde cross-linkers. Nature Communications, 13(1), 1468.