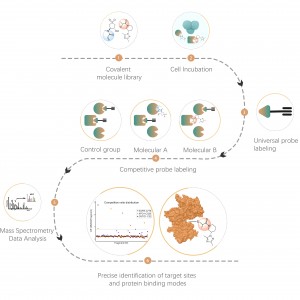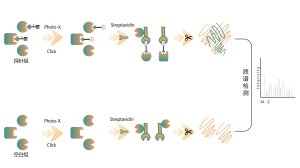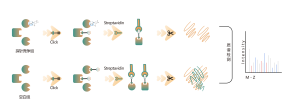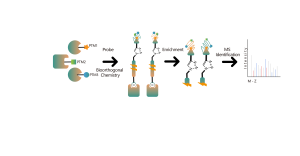
Products
Identification of Protein-Protein Interaction in Living Cells: Proximity Labeling
Technical Platform
The TurboID technology platform involves fusing a mutated biotin ligase to the C-terminus of a target protein. In overexpressed cells, the addition of biotin rapidly biotinylates interacting proteins within 10 minutes, even at room temperature, with minimal background noise. This versatility makes TurboID suitable for a wide range of biological systems, including mammalian cells, plants, and insects. Biotin-labeled proteins are then isolated, enriched, and digested to produce peptide samples for mass spectrometry, enabling detailed analysis of protein interactions and functions.

Our Advantages
01.High catalytic efficiency
02.Does not harm living cells
03.High biotinylation efficiency and stability
04.Effective biotinylation at room temperature (25°C)
Our Service
| Project | Identification of Protein-Protein Interaction Networks in Cells |
| Sample | Cell lysate, live cells |
| Hardware Platform | Non-contact ultrasonic cell pulverizer,ChemiDoc MP Imaging System,Orbitrap Fusion Lumos Tribrid/Orbitrap Exploris 480/Q Exactive HF-X/timsTOF Pro 2 mass spectrometer |
| Project Duration | 4-6 weeks |
| Deliverables | Project Report (including experimental procedures, data analysis charts, bioinformatics analysis results) |
| Price | Click to consult |
Case Study
Project Aim: Analyze changes in the protein interactome of the target protein (POI) in cells before and after drug treatment.
Solution: Utilize a proximity labeling strategy by constructing a POI-TurboID expression vector, overexpressing it in cells, and setting up drug-treated and control groups for biotinylation. Enrich the protein interactome in cells, followed by qualitative and quantitative analysis of differences in biotinylated proteins to characterize changes in the POI interactome.
Data visualization: