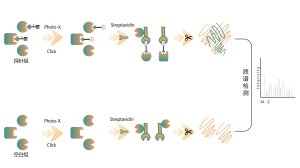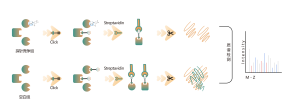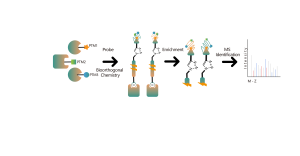
Products
Chemoproteomic discovery of New Lead Structures for undruggable targets
Technical Background
Currently, only ~800 proteins have been targeted by FDA-approved drugs, and a large number of disease-related targets are "undruggable". Because, at present, most technologies rely on purified proteins. The advent of chemproteomics has revolutionized drug discovery from purified proteins to living cells. It is capable of quantitatively analyzing the interactions between small molecules and proteins in the human proteome scale. Now the discovery of covalent structures against specific amino acid residues of protein targets, such as cysteine, lysine, methionine, and tyrosine, has been demonstrated in cell lysates and living cells. ChomiX will leverage its own chemoproteomics platform to break through the barriers of “undruggable” targets quickly.

Technical Advantages
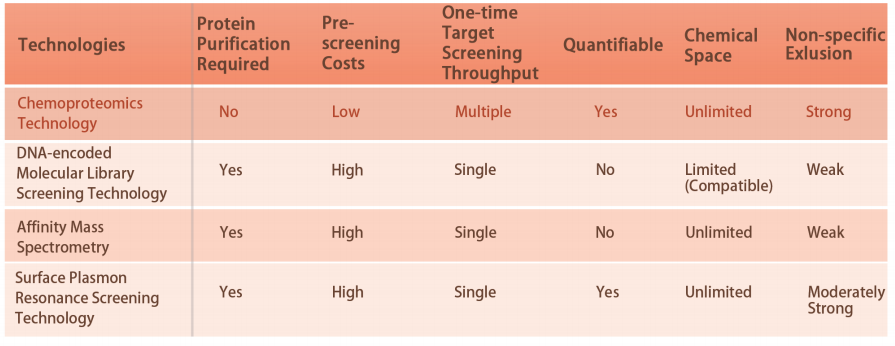
Platform Technical Features
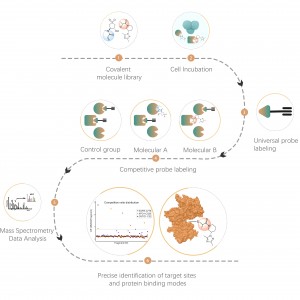
DIA-ABPP platform is designed to discover covalent binders that engage protein targets through the covalent reactions with the thiol group of cysteine residues. Once active cysteine residues of proteins are occupied by covalent molecules preferentially, the label of the universal probe that specifically reacts with the thiol group will be competed. In combination with DIA-based quantitative proteomics techniques, the intensities of probe-labeled peptides between covalent molecule and control group could be quantified. These signal differences represent the occupation rates of covalent molecules against their binding proteins. Unlike traditional single-target screening strategies, DIA-ABPP platform enables quantitative analysis of interactions between fragment electrophiles and nearly all proteins in living cells with the resolution of amino acid residue, which greatly improves efficiency and reduces risk for the high throughput lead structures discovery project.

Workflow of Covalent Lead Compound Screening
Feature 1

Our drug-like molecule library contains a variety of reactive groups (warheads) characterized by mild electrophilic reactivity such as acrylamide, chloroacetamide and alkynamide. These molecules feature structurally diverse pharmacophores that conform to Lipinski's Rule of Five.
Feature 2

Currently, the library of protein targets captured by the thiol-specific chemical probe covers 39962 cysteine sites from 12421 proteins, including kinase, phosphatase, ligases, and transcription factors. The library remains continuously updated, ensuring access to the latest information on specific targets and sites. Detailed information regarding specific targets and sites are available through the link below.
Feature 3

In a single screening experiment, the platform is able to quantify the target engagement of small molecules (A2-H6) against all labeled proteins in the whole proteome. The figure above shows site occupancy of selected clinically important targets by covalent molecules. The darker the color, the higher the occupancy of molecules to binding pockets, including orthosteric and allosteric sites (the asterisk indicates that the occupancy rate is higher than 80%). Thus, the affinity and selectivity of each molecule can be evaluated simultaneously for subsequent structure optimization.