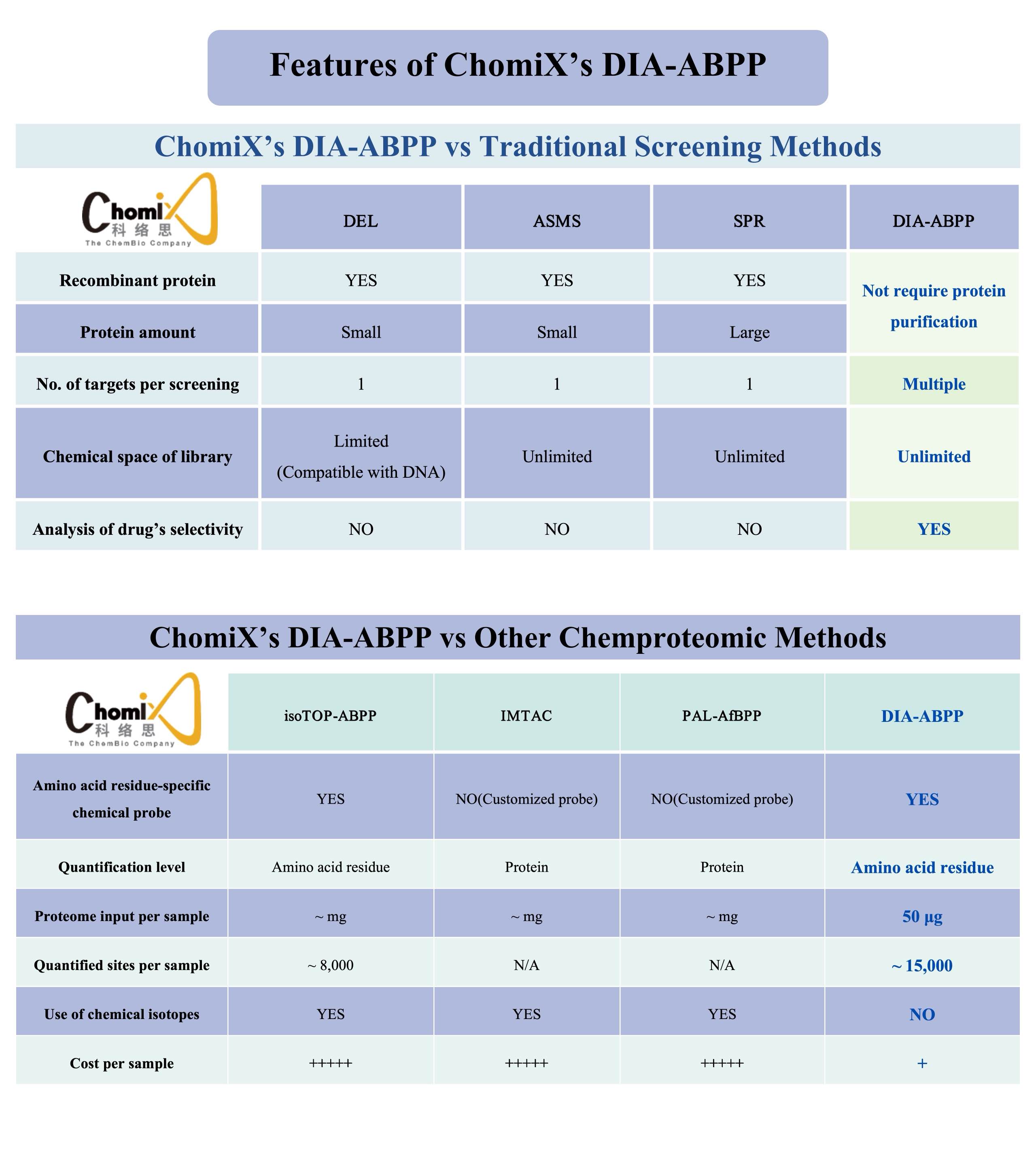
私たちの科学的プラットフォームは、ChomiXスクリーンは、DIA-ABPP (データに依存しない取得活性ベースのタンパク質プロファイリング) に基づいており、当社の ABPP をリードする方法論の重要な部分です。私たちのChomiXスクリーン技術プラットフォームを活用すれば、共有結合性リード化合物をスクリーニングして、創薬不可能な標的を見つけることができます。具体的なプロセスには、生細胞の共有結合分子処理、化学プローブで標識されたペプチドの分離と濃縮、高分解能質量分析による検出、薬物標的部位結合ネットワークの構築が含まれます。これにより、複数のタンパク質標的上の特定のアミノ酸残基ポケットに対する薬物の結合能と選択性を同時に評価し、候補化合物を生成することが可能になります。現在、システインを標的とした国内最大の共有結合性化合物ライブラリーを含む、包括的なタンパク質標的ライブラリーを構築しています。 、当社の化学プローブ ライブラリおよびその他の特徴的なコア モジュールに基づいています。
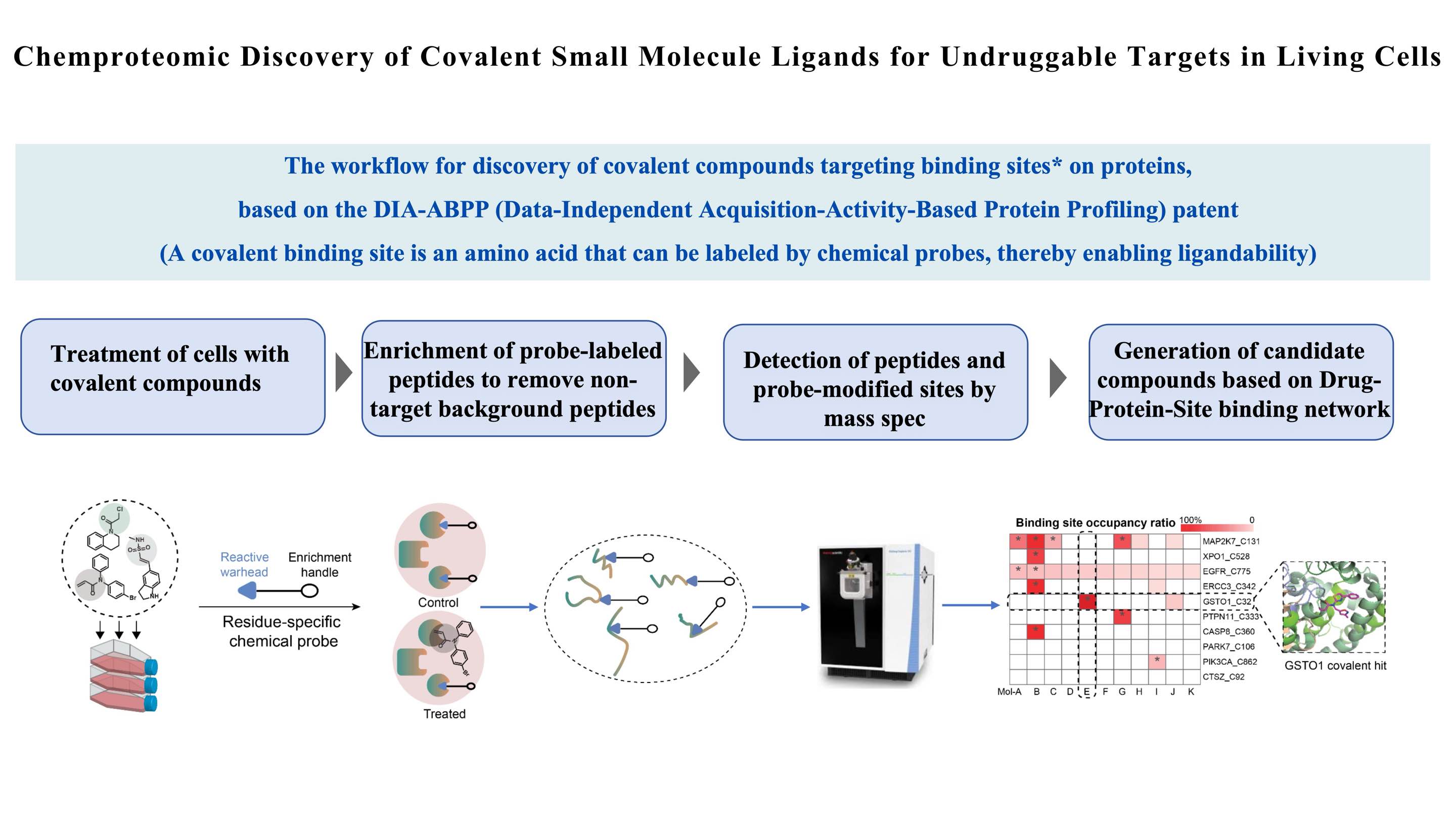
各化合物の結合部位 (Protein_Cys 部位) の占有率は、定量化されたプローブ標識ペプチドの質量スペクトル強度を使用して決定されます。本質的に、システイン特異的プローブは、特定の結合ポケットへの結合をめぐって化合物と競合します。プローブ標識ペプチドのシグナルが低いほど、化合物の占有能力は強くなります。対象タンパク質 (POI) の各システイン部位について、選択した化合物について用量依存性の標的結合率 (TE50) の決定が行われます。その後、TE50 値のランキングによってヒット候補が確認されます。 TE50 が低いほど、化合物が生細胞内の POI のシステイン部位をより強く占有することになります。