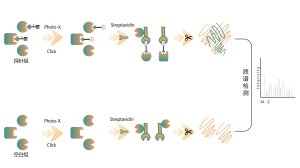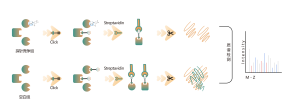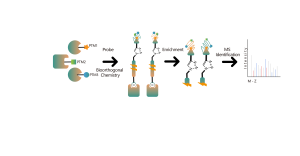
Products
Identification of Non-covalent Small Molecule Binding Pockets in Cells
Platform Technical Features
It is critical to determine the binding mode between small molecule drugs and their protein targets for drug R&D. A comprehensive analysis of these interactions at both structural and physico-chemical levels could significantly deepen our understanding of protein functions and facilitate drug design & optimization. Structural biology techniques, including X-ray, cryo-electron microscopy (cryo-EM), nuclear magnetic resonance (NMR), etc., have been widely used in the determination of drug-binding modes. The high-resolution structures of protein-drug complexes could greatly benefit the optimization of drug structures in the early stage. However, protein structure analysis has consistently posed challenges in life science research, especially for membrane protein targets like G protein-coupled receptors (GPCRs) and ion channel proteins. It often consumes substantial time and resources on processes such as protein purification, screening for protein-drug complex crystallization, and data acquisition & processing.
An ideal approach is to identify protein-drug interaction modes within living cells. This approach not only avoids high costs associated with structural studies, but also eliminate the possible false positives resulting from artificial conditions like high-salt buffers or saturated drugs.

Workflow

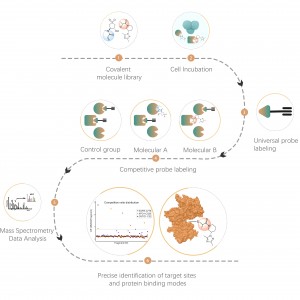
ChomiX employs the photoaffinity chemical probes derived from the non-covalent small molecule drugs, enabling the capture of labeled peptides located in binding pockets from living cells and subsequent identification by mass spectrometry. Once peptide sequences and labeled sites are determined, the exact binding mode could be rapidly obtained with the help of molecular docking.
Technical Advantages

Case Study
Project Aim
The putative target of Drug B is a transmembrane protein with multiple reported drug-binding pockets. Structural biological methods such as X-ray and cryo-EM failed. The chemoproteomic strategy will be tried to get the binding mode in living cells.
Experimental Method
The photoaffinity Probe B harboring the photo-crosslinking and bioorthogonal moieties was designed and synthesized. The target engagement of Drug B was firstly confirmed and the labeled peptide located in the binding pocket was sequenced by MS.
Data Visualization

Immunoblot and MS-based chemoproteomic data revealed that the drug candidate can effectively compete for the probe-labeled signal, indicating direct binding of the drug candidate to the target protein in living cells.

MS/MS spectrum for drug modification peptide: CLPFIIGCNPTILH*VHELYIR

Drug binding mode based on MS-based chemoproteomic data and molecular docking