Nucleic Acid-Protein Interactions Revealed Through Proximity Labeling Strategy: Exploring G4PID Probes and PLGPB Strategy
This article presents an innovative study wherein the authors introduce a novel bifunctional probe termed G4PID. This probe combines the G4-binding domain (RHAU23) of RHAU with the miniTurbo biotin ligase, enabling precise targeting of G-quadruplexes (G4) and tagging G4-interacting proteins within live cells. G4 structures, formed by guanine-rich nucleic acid sequences, are pivotal in various cellular processes including gene regulation, DNA replication, and repair. Despite widespread recognition of G4's significance, the specific mechanisms within cells and their interactions with proteins remain largely unexplored. Employing G4PID, the authors developed a specialized biotin labeling method (PLGPB) to accurately identify and analyze G4-interacting proteins, shedding light on the crucial role of G4 structures in cellular functions. This approach successfully identified 149 protein candidates interacting with G4, predominantly involved in transcriptional regulation, mRNA splicing, and chromatin remodeling. Validation of seven candidate proteins revealed their preference for RNA G4 binding, albeit with varying affinities towards DNA G4. Detailed examination of the splicing factor SF3B4 demonstrated its interaction with G4 structures, impacting alternative splicing events. Stabilization of G4 structures altered SF3B4's binding efficiency and influenced alternative splicing of specific genes (e.g., INPPL1 and PPP6R2).
Berberine, a traditional alkaloid with wide-ranging pharmacological effects including anti-inflammation, hypoglycemia, and cardiovascular protection, has drawn considerable attention. However, its precise molecular mechanisms, particularly in inflammation suppression, remain unclear.
This study fills this knowledge gap using ABPP technology to identify EIF2AK2 as a critical target engaged by berberine for its anti-inflammatory action. The findings deepen our understanding of berberine's mechanism and provide a scientific basis for repositioning berberine and developing new EIF2AK2-targeted anti-inflammatory drugs.
The team employed advanced chemoproteomic methods to systematically investigate berberine's interactions with intracellular proteins, confirming its specific binding to EIF2AK2 and modulation of its enzymatic activity. This influences inflammatory response pathways, effectively inhibiting inflammation progression. This significant breakthrough offers insights into berberine's anti-inflammatory mechanism and supports the development of novel therapies targeting EIF2AK2.
ChomiX provides cutting-edge services like ABPP and CETSA to aid researchers in exploring drug mechanisms and expediting new drug development efforts.
Research Route
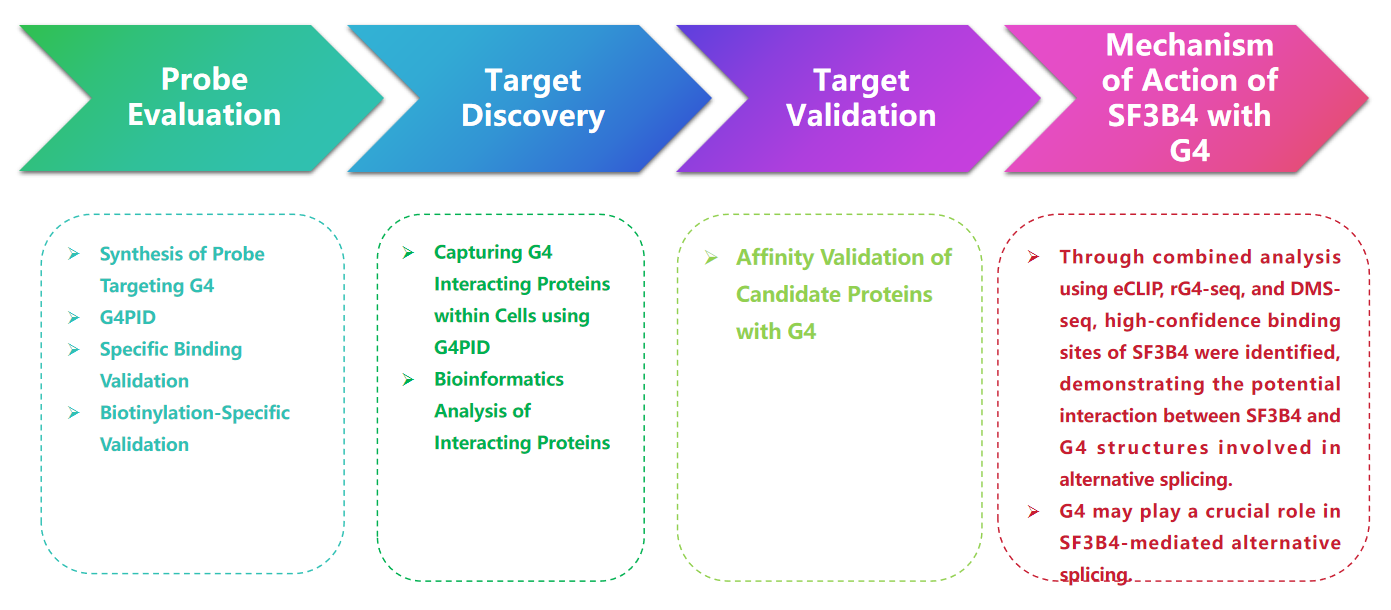
Experimental process
1. Construction and Specificity Verification of the G4PID Probe.
The authors initially engineered the G4PID probe, with the G4-binding domain RHAU23 positioned at the N-terminus and the miniTurbo at the C-terminus, linked by a flexible linker. Expression and purification were carried out using the BL21 (DE3) strain. Specific binding validation experiments with purified G4PID revealed its precise binding to BCL2 G4, exhibiting a Kd value of 15 ± 7 nM, while displaying negligible affinity towards other non-G4 structures. Furthermore, the authors assessed the biotinylation specificity of G4PID, demonstrating its capability to selectively biotinylate G4-interacting proteins.
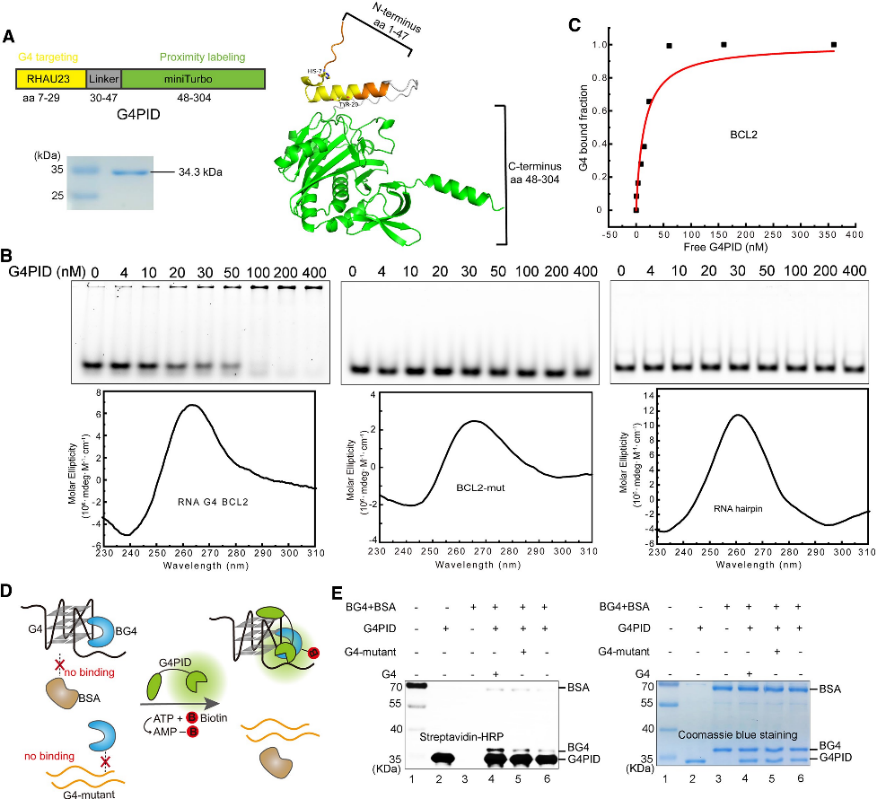
Figure 1 G4 PID mediates the specific biotinylation of the G 4-interacting protein
2. RNA G4 Binding Capability of G4PID in Cells and Biotin Labeling Characteristics.
The authors established HA-G4PID and HA-miniTurbo HeLa cell lines to investigate G4PID's binding affinity to intracellular RNA G4 via eCLIP experiments. Results demonstrate the selective interaction of G4PID with RNA G4, exhibiting notably higher binding affinity to RNA sequences containing PQS compared to random sequences. Additionally, the authors observed unique features of G4PID facilitating the initiation of biotin labeling within cells without requiring cofactors. This led to a significant enhancement in protein labeling efficiency, with evident biotinylation signals detected for G4PID within 30 minutes, indicating faster labeling kinetics compared to miniTurbo.
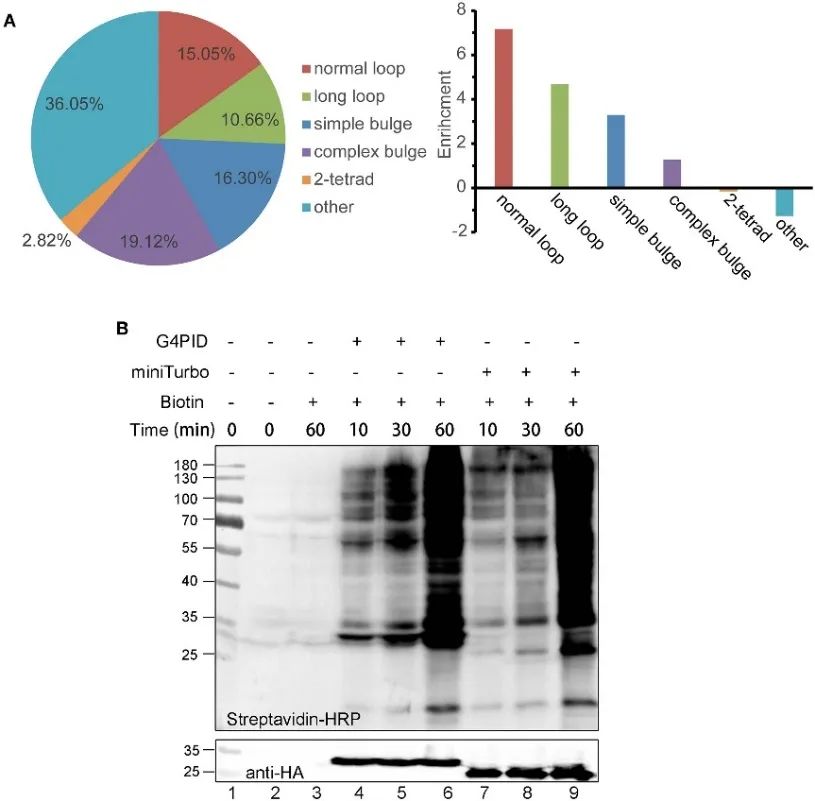
Figure 2 G 4 PID combined with RNA G4 experiments and optimization for the labeling efficiency in HEK293T cells.
3. Capturing and Detecting the Intracellular G4 Interacting Proteome Using the PLGPB Method.
Subsequently, the authors employed the G4PID probe to capture and detect the G4 interacting proteome within live cells using the Proximity Labeling of G4-Interacting Proteins (PLGPB) approach. In three independent replicate experiments encompassing 578 proteins, achieving an 80% identity, transfection with G4PID and miniTurbo in HEK293T cells was conducted. Screening revealed 149 candidate proteins interacting with G4, exhibiting significant overlap with existing databases of G4-interacting proteins. Furthermore, Gene Ontology (GO) enrichment analysis indicated a substantial association of these proteins with transcriptional regulation, mRNA splicing, and chromatin remodeling, consistent with previous findings in G4 research.
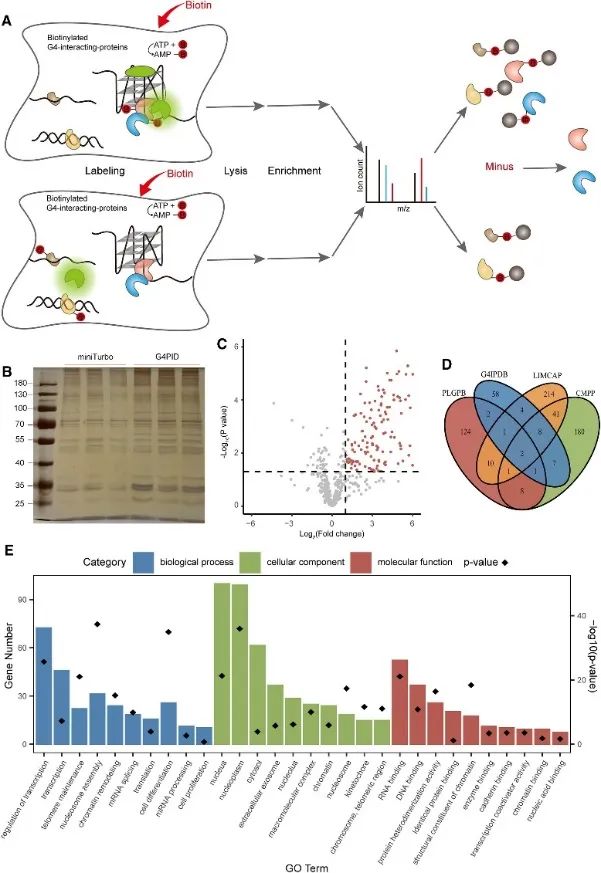
Figure 3 identified the proteins interacting with G4 by the PLGPB method.
4. Validation of G4 Binding Affinity in Newly Identified Candidates.
Through the PLGPB method, the authors discovered a range of candidates encompassing various functional categories, including common telomere-associated proteins, transcription factors, and relatively uncommon protein-binding proteins. This indicates that the PLGPB method not only reaffirms previously acknowledged G4 interacting proteins but also significantly broadens the protein landscape by uncovering novel candidates spanning diverse functional classes. Furthermore, the authors validated the binding capacity of these candidates to RNA G4 and DNA G4, revealing a pronounced preference for RNA G4 and variations in affinity towards DNA G4 in different conformations. These findings underscore the potential to modulate protein-G4 interaction by regulating G4 conformation.
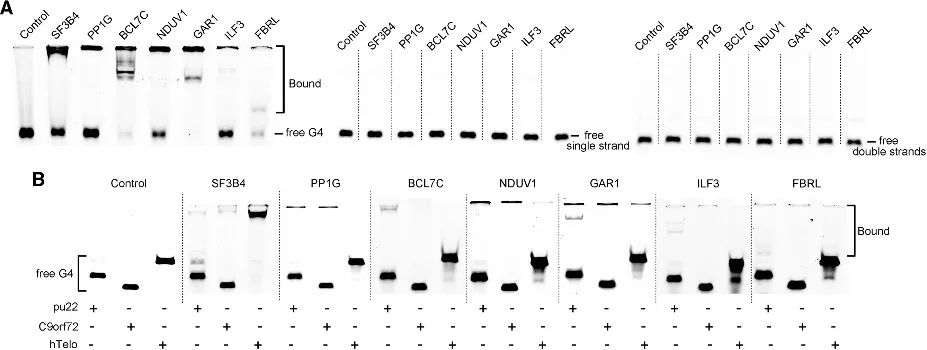
Figure 4 Interaction between the candidate proteins and RNA G4 and DNA G4.
5. Determining High Confidence Binding Sites of SF3B4 and G4 Structure.
During the investigation into the in vivo mode of G4 binding, the authors unearthed the significant role of the SF3B4 protein. SF3B4, known for its involvement in RNA splicing, had its high confidence binding sites identified through eCLIP experiments and a comprehensive analysis of rG4-seq and DMS-seq data. Approximately half of these binding sites contained PQS, predominantly associated with unconventional G4 structures. Moreover, the authors observed a close spatial relationship between SF3B4 binding sites and RNA PQS, with an enriched distribution primarily within intronic regions, particularly proximal to the 3' splice site. These findings strongly suggest that the interaction between SF3B4 and the G4 structure might play a crucial role in alternative splicing mechanisms.
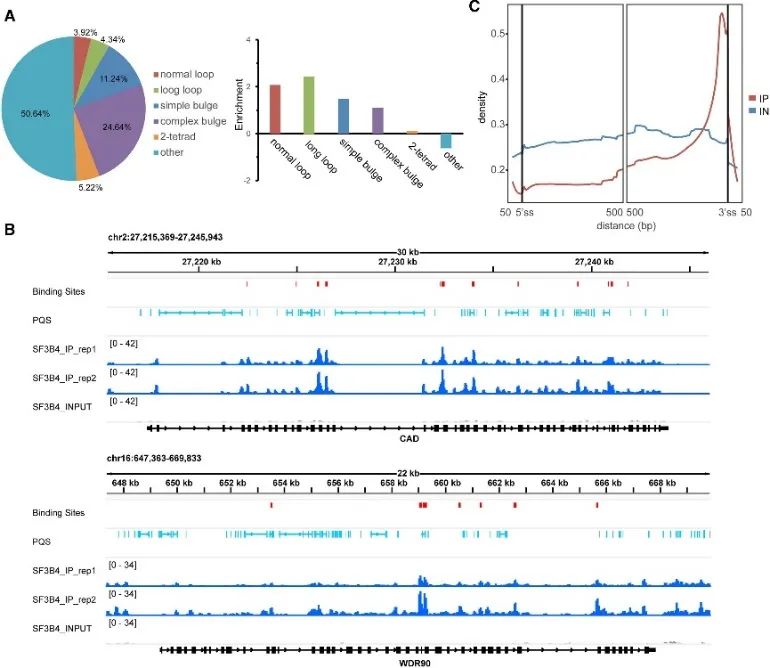
Figure 5 identifies the G4 binding preference of SF3B4 in cells using eCLIP technology.
6. G4 Structure Regulates SF3B4-Mediated Alternative Splicing.
The authors conducted an analysis of SF3B4 binding sites to unveil its association with the G4 structure and further investigated its regulatory mechanism in alternative splicing events. Conventional PQS implicated in alternative splicing were screened, with two genes, INPPL1 and PPP6R2, selected for detailed examination. G4 structures within INPPL1 and PPP6R2, termed G4-INPPL1 and G4-PPP6R2, were confirmed, and SF3B4 was identified as specifically binding to these G4 structures. Furthermore, it was demonstrated that the addition of the G4 ligand pyridostatin (PDS) diminishes SF3B4 binding to G4 structures, consequently promoting exon inclusion events during INPPL1 and PPP6R2 mRNA splicing. These findings indicate the potential significance of the G4 structure in SF3B4-mediated alternative splicing.
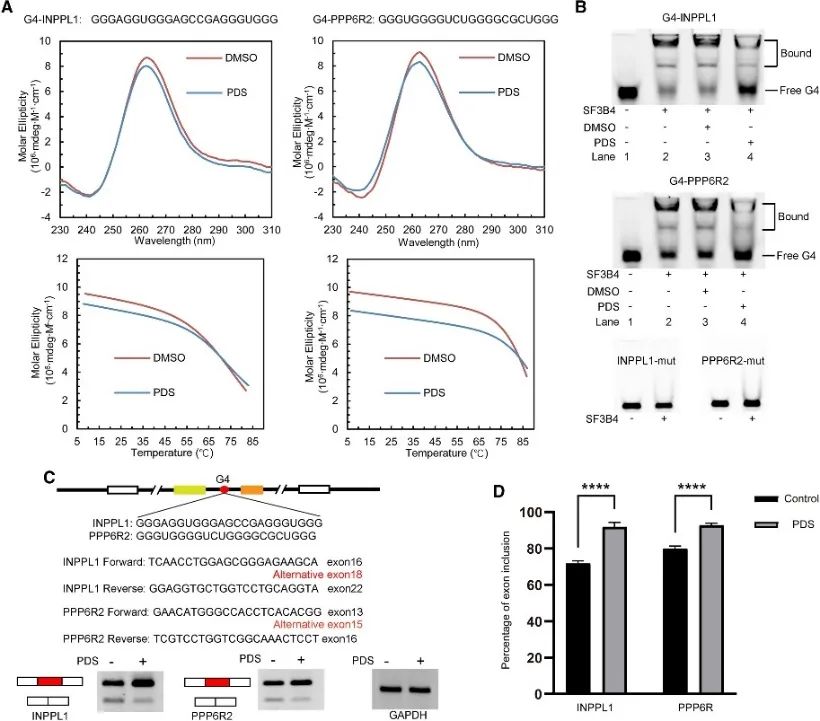
Figure 6 SF3B4 binds the G4 structure and affects the alternative splicing of the mRNA.
In conclusion, this study introduces an efficient toolkit comprising the G4PID probe and PLGPB strategy, shedding light on how the interplay between G4 and proteins within the cell impacts gene expression, particularly influencing the pivotal process of alternative splicing. These findings deepen our comprehension of the regulatory mechanisms involving G4 in cellular biological processes and pave the way for further exploration into therapeutic strategies for G4-related diseases.