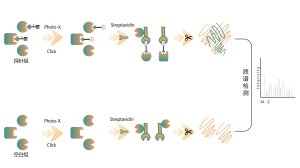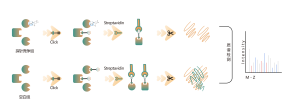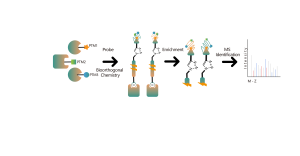
Products
Protein Post-translational Modifications (PTMs) Analysis Based on Chemical Probes
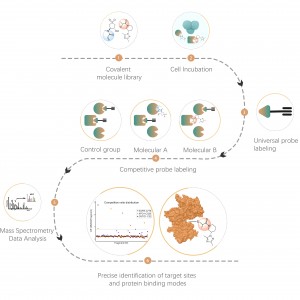
In addition to traditional antibody enrichment strategies, chemical, enzymatic, and metabolic labeling techniques have been progressively applied in the enrichment and identification of post-translational modifications (PTM) in proteins. The underlying principle involves using chemical reaction or enzyme catalysis to covalently attach specific molecular probes to target modifications or modifying residues, followed by proteomic analysis. This emerging approach in chemoproteomics is becoming important in the study of low-abundance protein modifications, such as palmitoylation, myristoylation and glycosylation. Notably, it offers the advantage of being applicable in living cells, thereby circumventing potential interference, such as oxidation resulting from cell stress.

Technical Service
1. Cysteine Post-Translational Modifications
Cysteine (Cys) is a common sulfur-containing amino acid in proteins, and its thiol group plays crucial roles in regulating molecular redox homeostasis, influencing enzymatic reactions, protein-protein interactions, and protein stability through diverse post-translational modification processes. Common post-translational modifications on cysteine include palmitoylation, nitrosylation, sulfenylation, and sulfonation. Colosseum Biosciences has developed universal probes or metabolic labeling strategies targeting cysteine residues and established corresponding chemical proteomics platforms for post-translational modification identification. This platform accurately identifies target proteins and modification sites of post-translational modifications.
2. Lysine Post-Translational Modifications
Lysine (Lys) is one of the amino acids with the most diverse post-translational modifications (PTMs) in the proteome. Reversible lysine post-translational modifications (Lys-PTMs) are common biochemical processes that play indispensable roles in regulating numerous critical cellular functions. Post-translational modifications on lysine include acetylation, succinylation, propionylation, butyrylation, crotonylation, malonylation, and lactylation. Colosseum Biosciences has developed a chemical proteomics technology platform based on chemical probes for lysine post-translational modifications, which accurately identifies target proteins and modification sites.
3. Serine/Threonine Post-Translational Modifications
Common post-translational modifications on serine (Ser) and threonine (Thr) mainly include phosphorylation, O-glycosylation, etc. These modifications significantly impact the activity state of proteins and their interactions with other molecules, playing important roles in various key biological processes. Therefore, the analysis of this type of post-translational modification is an important topic in protein function research. Colosseum Biosciences utilizes phosphorylation enrichment materials, non-natural glycan chemical probes, and other tools to accurately identify target proteins and modification sites for this type of post-translational modification.
Our Advantages
01. Highly Professional: Powered by the research group of Professor Wang Chu from Peking University, the chief scientist of our company.
02. One-stop Platform: Encompassing the entire workflow from probe design, probe synthesis, target discovery, to bioinformatics analysis.
03. No Need for antibodies: Specific chemical probe modification, Physiological
04. Environment: Applicable in both lysates and living cells.
05. Extensive Experience: Accumulated expertise in over 10 different types of PTMs
06. High-performance Platform: Thermo Fisher Orbitrap Exploris 480, Thermo Fisher Q Exactive HF-X, Bruker timsTOF
Case Study
Itaconation refers to the process in which itaconate, covalently binds to specific sites on proteins during post-translational modification. This modification can alter the protein's structure and function, influencing its role within the cell. For example, itaconation can regulate enzyme activity, stability, or interactions with other molecules. Since itaconate is an intermediate in the tricarboxylic acid cycle, this modification is closely linked to the cell's metabolic state. Research has shown that itaconation plays a significant role in the pathology of certain diseases, and understanding this modification mechanism could aid in developing new therapeutic strategies.
The cells metabolically labeled with ITalk probes at the living cell level were lysed, followed by click reaction, enrichment, enzyme digestion and other steps to determine the specific site of itaconic acidification.