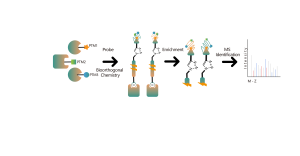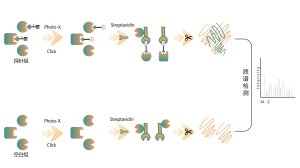
Products
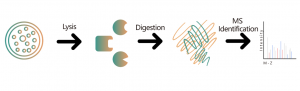
Untargeted Protein Qualitative/Quantitative Analysis
In the field of drug development, scientists have been dedicated to exploring innovative therapies for specific diseases. Differential protein analysis has become a key tool for gaining insight into disease molecular mechanisms, identifying effective therapeutic targets, and providing critical clues and scientific evidence for the discovery and development of novel drugs. This technology empowers researchers to systematically detect changes in protein expression, discover disease-related target proteins, and guide the design of new drug development and personalized treatment strategies.
With the development and application of cutting-edge technologies such as high-throughput sequencing and mass spectrometry, the accuracy and coverage of differential protein analysis have significantly improved. This advancement enables researchers to delve deeply into and meticulously evaluate potential drug targets in disease progression based on large-scale data. Therefore, in modern biomedical research, differential protein analysis is not only a key approach to unraveling the complex biological processes of diseases but also an essential engine driving the progress of new drug development.
Proteomic Quantification Methods

Our Advantages
1. Professional Excellence: Our team boasts extensive experience and publications in top journals, offering industry-leading technical services.
2. Efficient Solutions: We employ reliable methods to drive projects forward swiftly, providing worry-free solutions.
3. Rigorous Quality Management: Adhering to ISO 9001 standards, our mature quality management system ensures the authenticity and reliability of our reports.
4. Systematic Project Management: From consultation to report delivery, we provide timely progress updates, ensuring customer satisfaction and efficient project execution.
5. Cutting-Edge Equipment: Equipped with advanced mass spectrometers like the Thermo Fisher Orbitrap Exploris 480 and Bruker timsTOF, we facilitate groundbreaking research.
Our Service
| Project | Qualitative/Quantitative Proteomics Analysis |
| Sample | Tissue, cell precipitate, lysate, purified protein |
| Hardware Platform | VanquishNeo UPLC coupled with Orbitrap Exploris 480 mass spectrometer (Thermo Fisher Scientific); EASY-nLC1200 UPLC coupled with Q Exactive HF-X mass spectrometer (Thermo Fisher Scientific) |
| Project Duration | 4-8 weeks |
| Deliverables | Project Report (including lists of qualitatively/quantitatively identified proteins, bioinformatics analysis, quality control analysis, etc.) |
| Price | Click to consult |
Case Study
Project Introduction:Comparative analysis of changes in whole proteome levels between the drug-treated group and control group to investigate molecular mechanisms underlying drug phenotype.
Sample Types:Cellular specimens subjected to drug and control treatments, each comprising three biological replicates.
Experimental Method:Quantitative identification of differentially expressed proteins at the whole proteome level using TMT-based multiple isotope labeling proteomics methodology.

1. As shown in the volcano plot of differential protein abundance, a total of 5,987 proteins were quantified across all six sample groups. Statistical testing was conducted on the ratio of each protein. In the drug-treated group, 560 proteins exhibited upregulation, while 363 proteins showed downregulation in abundance. The corresponding intensity information was also visualized using heatmaps.
2. KEGG pathway and Gene Ontology (GO) analyses were conducted on the differentially expressed proteins, including GOTERM_Biological Process, GOTERM_Cellular Component, and GOTERM_Molecular Function. By assessing the significance level of GO term enrichment, we identified functional categories and pathways significantly enriched by the differentially expressed proteins, thereby contributing to the exploration of drug molecular mechanisms.
3. Exemplified by the above figure, significantly enriched upregulated proteins were observed in signaling pathways such as nuclear chromosome segregation, mitotic sister chromatid segregation, and sister chromatid segregation. This indicates that the drug, at the molecular level, influences the process of chromatin separation within the nucleus.