-
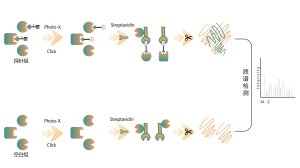
Chemoproteomic profiling of protein targets for noncovalent small molecule drugs
Non-covalent interactions, such as hydrogen bonds and π-π stacking, can be disrupted due to protein denaturation. To address this challenge, ChomiX platform employs photoaffinity labeling, a well-established technique for precisely attaching “chemical labels” to a protein’s active site. Furthermore, ChomiX’s innovative in situ chemical crosslinking strategy transforms transient non-covalent protein interactions into covalent and permanent chemical bonds. By utilizing a chemical probe that is functionalized with both photoaffinity and bioorthogonal moieties, ChomiX’s chemoproteomics platform has demonstrated its effectiveness in successfully fishing out protein targets within cell lysates, tissues, and living cells. The spectrum of bioactive small molecule drugs applied in the platform encompasses a variety of compounds, including endogenous metabolites, natural products, and non-covalent synthetic molecules.
-
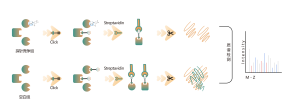
Competitive chemoproteomic profiling of protein targets for covalent small molecule drugs
Similar to non-covalent drugs, the direct pulldown strategy by using chemical probes functionalized with reporter groups have also been demonstrated successfully for covalent small molecule drugs. However, it is worth noting that some covalent drugs are intolerant of chemical modifications due to loss of bioactivity or synthetic inaccessibility. In addition, the formed covalent bond is usually unstable during MS detection.
Competitive chemoproteomic strategy is an ideal alternative, which uses a universal activity-based probe for protein labeling. Specific chemical probes have developed to react with cysteine, lysine, serine, and histidine residues. In principle, once occupied by a covalent small molecule, the amino acid residue cannot be labelled by the probe. As a result, the ON and OFF-targets could be identified comprehensively by this competitive strategy with the resolution of amino acid.
-
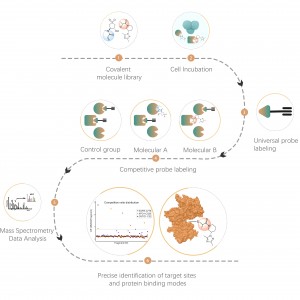
Chemoproteomic Discovery of New Lead Structures for Undruggable Targets
Chemoproteomic discovery of New Lead Structures for undruggable targets Technical Background Currently, only ~800 proteins have been targeted by FDA-approved drugs, and a large number of disease-related targets are ”undruggable“. Because, at present, most technologies rely on purified proteins. The advent of chemproteomics has revolutionized drug discovery from purified proteins to living cells. It is capable of quantitatively analyzing the interactions between small molecules and...